13.11 Creating a Dendrogram
13.11.2 Solution
Use hclust() and plot the output from it. This can require a fair bit of data preprocessing. For this example, we’ll first take a subset of the countries data set from the year 2009. For simplicity, we’ll also drop all rows that contain an NA, and then select a random 25 of the remaining rows:
library(dplyr)
library(tidyr) # For drop_na function
library(gcookbook) # For the data set
# Set random seed to make random operation below repeatable
set.seed(392)
c2 <- countries %>%
filter(Year == 2009) %>% # Get data from year 2009
drop_na() %>% # Drop rows that have any NA values
sample_n(25) # Select 25 random rows
c2
#> Name Code Year GDP laborrate healthexp infmortality
#> 1 Egypt, Arab Rep. EGY 2009 2370.7111 48.8 113.29717 20.0
#> 2 Haiti HTI 2009 656.7792 69.9 39.60249 59.3
#> 3 Belize BLZ 2009 4056.1224 64.1 217.15214 14.9
#> ...<19 more rows>...
#> 23 Luxembourg LUX 2009 106252.2442 55.5 8182.85511 2.2
#> 24 Lithuania LTU 2009 11033.5885 55.7 729.78492 5.7
#> 25 Denmark DNK 2009 55933.3545 65.4 6272.72868 3.4Notice that the row names (the first column) are essentially random numbers, since the rows were selected randomly. We need to do a few more things to the data before making a dendrogram from it. First, we need to set the row names-right now there’s a column called Name, but the row names are those random numbers (we don’t often use row names, but for the hclust() function they’re essential). Next, we’ll need to drop all the columns that aren’t values used for clustering. These columns are Name, Code, and Year:
rownames(c2) <- c2$Name
c2 <- c2[, 4:7]
c2
#> GDP laborrate healthexp infmortality
#> Egypt, Arab Rep. 2370.7111 48.8 113.29717 20.0
#> Haiti 656.7792 69.9 39.60249 59.3
#> Belize 4056.1224 64.1 217.15214 14.9
#> ...<19 more rows>...
#> Luxembourg 106252.2442 55.5 8182.85511 2.2
#> Lithuania 11033.5885 55.7 729.78492 5.7
#> Denmark 55933.3545 65.4 6272.72868 3.4The values for GDP are several orders of magnitude larger than the values for, say, infmortality. Because of this, the effect of infmortality on the clustering will be negligible compared to the effect of GDP. This probably isn’t what we want. To address this issue, we’ll scale the data:
c3 <- scale(c2)
c3
#> GDP laborrate healthexp infmortality
#> Egypt, Arab Rep. -0.6183699 -1.564089832 -0.6463085 -0.2387436
#> Haiti -0.6882421 0.602552464 -0.6797433 1.0985742
#> Belize -0.5496604 0.006982544 -0.5991902 -0.4122886
#> ...<19 more rows>...
#> Luxembourg 3.6165896 -0.876103889 3.0147969 -0.8444498
#> Lithuania -0.2652086 -0.855566996 -0.3666121 -0.7253503
#> Denmark 1.5652292 0.140472354 2.1481851 -0.8036157By default the scale() function scales each column relative to its standard deviation, but other methods may be used.
Finally, we’re ready to make the dendrogram, as shown in Figure 13.19:
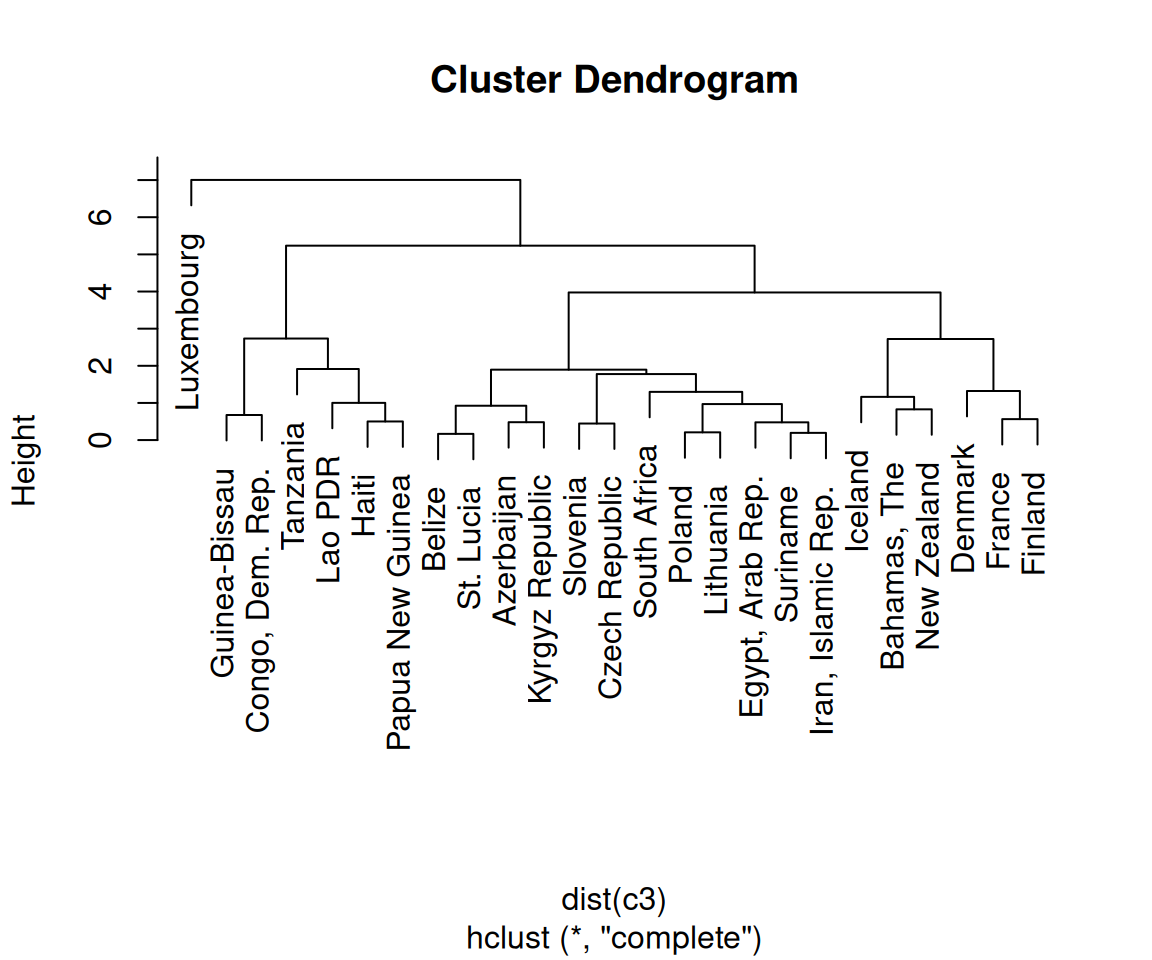
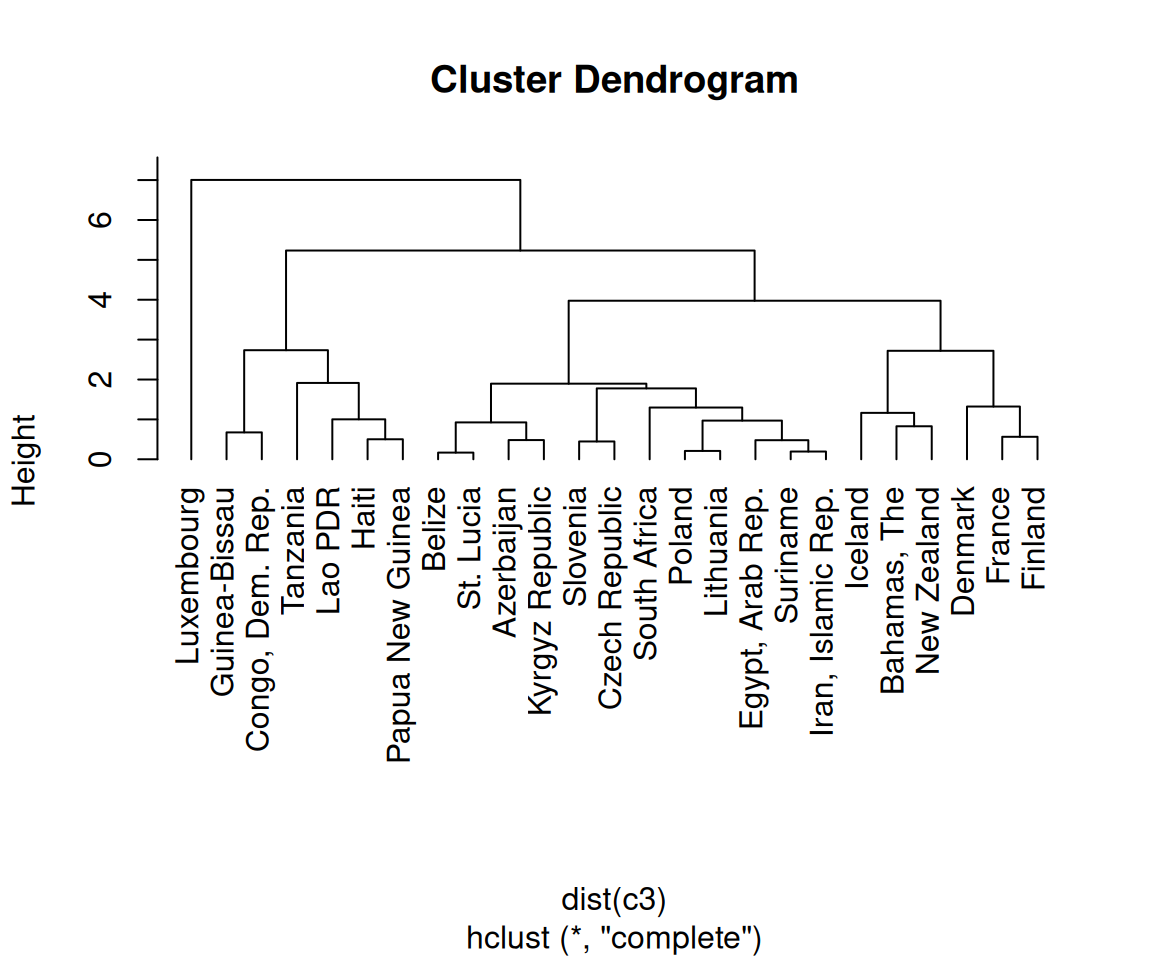
Figure 13.19: A dendrogram (left); With text aligned (right)
13.11.3 Discussion
A cluster analysis is simply a way of assigning points to groups in an n-dimensional space (four dimensions, in this example). A hierarchical cluster analysis divides each group into two smaller groups, and can be represented with the dendrograms in this recipe. There are many different parameters you can control in the hierarchical cluster analysis process, and there may not be a single “right” way to do it for your data.
First, we normalized the data using scale() with its default settings. You can scale your data differently, or not at all. (With this data set, not scaling the data will lead to GDP overwhelming the other variables, as shown in Figure 13.20.)
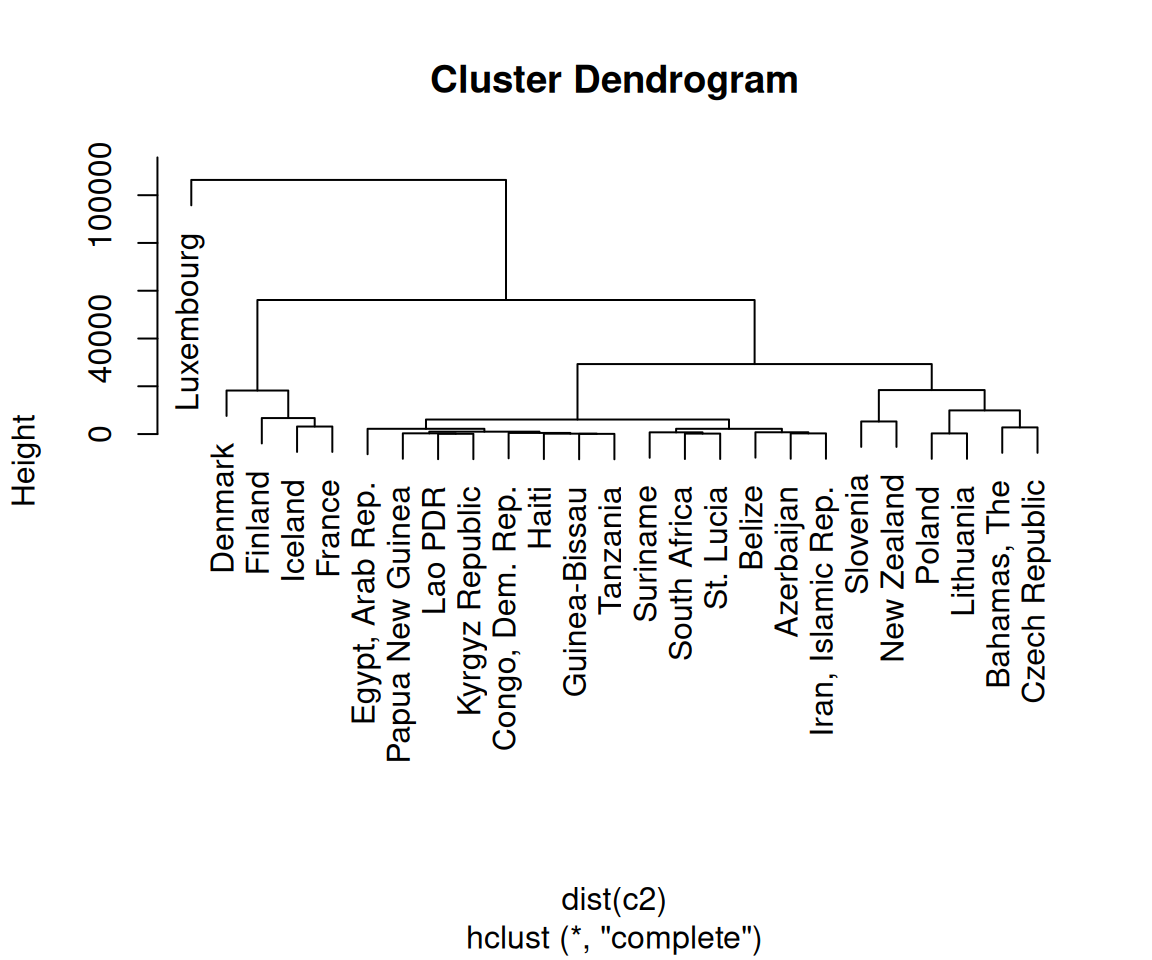
Figure 13.20: Dendrogram with unscaled data-notice the much larger Height values, which are largely due to the unscaled GDP values
For the distance calculation, we used the default method, “euclidean”, which calculates the Euclidean distance between the points. The other possible methods are “maximum”, “manhattan”, “canberra”, “binary”, and “minkowski”.
The hclust() function provides several methods for performing the cluster analysis. The default is “complete”; the other possible methods are “ward”, “single”, “average”, “mcquitty”, “median”, and “centroid”.